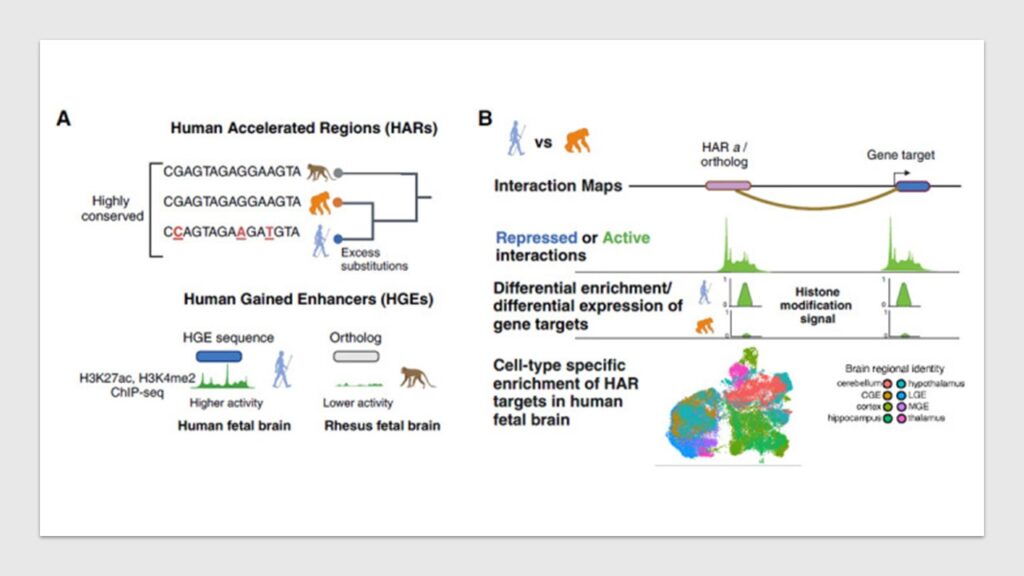
The human brain is a marvel of evolution, exhibiting unique features such as an increased cortical surface area, prolonged neurogenesis, and advanced synaptic plasticity. Scientists have long sought to unravel the genetic factors behind these developments. One crucial discovery in this domain is the presence of Human Accelerated Regions (HARs)—stretches of DNA that have remained highly conserved across species but show rapid evolution in humans. HARs are believed to function as transcriptional enhancers, regulating genes involved in neurodevelopment. Yet, the precise genes and pathways they influence have remained largely unknown.
A recent research by Yale University published on Cell sheds new light on this mystery by mapping chromatin interactions of HARs and their orthologs in human and chimpanzee neural stem cells (NSCs). The study explores whether HARs act by altering the expression of existing gene targets or by acquiring new gene targets—a mechanism known as enhancer hijacking. The results indicate that HARs predominantly regulate a conserved set of genes, many of which are implicated in neurodevelopment, rather than acquiring new targets.
Mapping the Interactions of HARs
To uncover the role of HARs, researchers employed Capture Hi-C (CHi-C), a technique that enables high-resolution mapping of chromatin interactions. This method was applied to 1,590 HARs and 466 Human Gained Enhancers (HGEs), another class of regulatory elements exhibiting human-specific activity. The study compared gene targets between human and chimpanzee NSCs to determine whether HARs influence species-specific or conserved genes.
Findings revealed that 88.9% of HARs and 75.8% of HGEs in human NSCs had identifiable gene targets. Notably, 2,963 genes were conserved across both species, with significant enrichment in neurodevelopmental functions such as axon guidance, synaptic transmission, and neuron migration. Conversely, species-specific HAR gene targets did not show significant enrichment for any biological function or differential gene expression, suggesting that HARs predominantly operate within conserved pathways rather than through enhancer hijacking.
Linking HARs to Neurodevelopmental Genes
A key question was whether HARs directly regulate the genes they interact with. To address this, the researchers used CRISPR activation (CRISPRa) and CRISPR interference (CRISPRi) to respectively enhance or repress the activity of HARs and their chimpanzee orthologs. One example, HACNS52, was found to regulate genes ANXA2 and ICE2, both of which have enriched expression in progenitor zones of the human fetal brain. Experimental results confirmed that repressing HACNS52 led to decreased expression of these genes, while activation resulted in increased expression.
Further analysis revealed that HAR-targeted genes were significantly overrepresented among genes showing human-specific expression changes in the developing and adult brain. This suggests that HARs contribute to unique human brain features by fine-tuning the expression of genes already present in primate neurodevelopmental pathways.
HARs and Their Role in Enhancer Activation
An unexpected finding was that some HARs and HGEs establish regulatory interactions in a repressed state in NSCs but activate upon neuronal differentiation. For instance, HAR116, which interacts with the gene TCF20, transitions from a repressed state in NSCs to an active state in neurons, corresponding with an increase in TCF20 expression. This indicates that HARs may act as developmental switches, influencing gene expression at critical stages of brain development.
The Cellular Context: HARs in Neural Subtypes
To understand the cellular environments in which HARs function, the researchers integrated their data with single-cell expression atlases of the developing human brain. They found that many HAR-regulated genes were preferentially expressed in outer radial glia (oRG), a neural progenitor population hypothesized to play a key role in cortical expansion. Genes such as ANXA2, ROBO1, and PLAGL1 showed a strong expression bias in oRGs, further reinforcing the idea that HARs contribute to the unique architecture of the human brain.
Addressing the Enhancer Hijacking Hypothesis
A major debate in evolutionary genetics is whether HARs contribute to human-specific traits through enhancer hijacking, wherein they acquire new gene targets that were not present in ancestral species. This study found little evidence to support this theory. Instead, HARs predominantly regulate genes that are already targeted by their chimpanzee orthologs. This suggests a conservative model of brain evolution, where uniquely human traits emerge through modifications of existing gene regulatory networks rather than through large-scale rewiring.
Implications for Understanding Neurodevelopmental Disorders
Interestingly, the study also found that HAR gene targets were significantly enriched in genes associated with autism spectrum disorder and schizophrenia. This raises the possibility that genetic variations in HARs could contribute to the risk of these conditions by disrupting normal gene regulatory mechanisms. Future studies could investigate whether HAR mutations correlate with neurodevelopmental disorders, providing new insights into their genetic underpinnings.
Conclusion: A Roadmap for Future Research
This research marks a significant step forward in our understanding of how human-specific genetic elements shape brain evolution. By establishing a high-resolution map of HAR interactions, the study provides a foundation for hypothesis-driven investigations into the functional roles of HARs.
Future research directions include studying HARs in more complex models such as cerebral organoids or humanized mouse models, investigating combinatorial effects of multiple HARs, as individual HARs may have subtle effects that become significant in concert, and exploring other developmental contexts beyond the brain, such as limb development, where HARs may have also played a role.


Leave a Reply