In an era where antibiotic resistance threatens to undermine modern medicine, researchers have made a groundbreaking discovery that could revitalize our ability to treat deadly bacterial infections. A recent study by Wang et al. reveals how tiny cellular messengers can silence the genes that make bacteria resistant to antibiotics, potentially providing a powerful new weapon in our fight against superbugs.
The problem: superbugs on the rise
The emergence of multidrug-resistant bacteria has become one of the most pressing global health challenges of our time. These “superbugs” have evolved mechanisms to survive even our strongest antibiotics, leaving clinicians with fewer treatment options for potentially fatal infections.
One particularly concerning pathogen is Methicillin-Resistant Staphylococcus aureus (MRSA), which has developed resistance to beta-lactam antibiotics through a gene called mecA. This gene produces a protein known as penicillin-binding protein 2a (PBP2a), which allows the bacteria to survive methicillin treatment. Traditional antibiotic development cannot keep pace with the rapid evolution of bacterial resistance, creating an urgent need for alternative therapeutic approaches.
The breakthrough: Silencing resistance genes
The research team discovered that tiny bubble-like structures called exosomes—naturally released by human cells as a way to transport materials between cells—that can be loaded with small interfering RNA (siRNA) and delivered into bacterial cells. These siRNAs are designed to target and silence specific bacterial genes. Until now, researchers could not achieve RNA interference (RNAi) in bacteria because these microorganisms lack the necessary cellular machinery. However, the innovation presented in this study demonstrates that human exosomes can transport not only siRNA, but also the necessary molecular machinery — including a crucial protein called Argonaute 2 (AGO2) — directly into bacterial cells, thereby enabling RNAi to occur even in these cells.
How the technology works
The researchers followed a systematic approach to develop and validate their technology.
Firstly, they engineered human cells to produce exosomes containing siRNA designed to target specific bacterial resistance genes. These exosomes efficiently transported their cargo into bacterial cells, with each bacterium receiving approximately 56-105 copies of siRNA. Subsequently, the exosomal AGO2 protein forms a complex with siRNA, binding to bacterial messenger RNA and preventing protein synthesis without destroying the mRNA—a process called translational repression.
Lastly, by specifically targeting the mecA gene in MRSA, the researchers were able to reduce PBP2a protein expression and restore the bacteria’s susceptibility to methicillin. The researchers made an important discovery during this process: unlike in human cells, where perfectly matched siRNAs typically cause mRNA degradation, in bacteria, the siRNA-AGO2 complex primarily blocks translation without breaking down the target mRNA. This represents a novel mechanism of gene silencing.
Putting it to the test: laboratory experiments
The research team conducted extensive testing to verify their approach by treating MRSA with siMecA-Exos (exosomes containing siRNA targeting mecA) in laboratory cultures. The treated MRSA showed significantly decreased PBP2a protein levels, a two-fold reduction in the minimum inhibitory concentration (MIC) of methicillin, and an enhanced killing when treated with a combination of siMecA-Exos and methicillin. The researchers critically confirmed that the exosomal AGO2 protein is essential for this gene silencing effect: in fact, when they prepared exosomes from cells with reduced AGO2 levels, the therapeutic effect was largely lost.
From lab to living organisms: mouse models
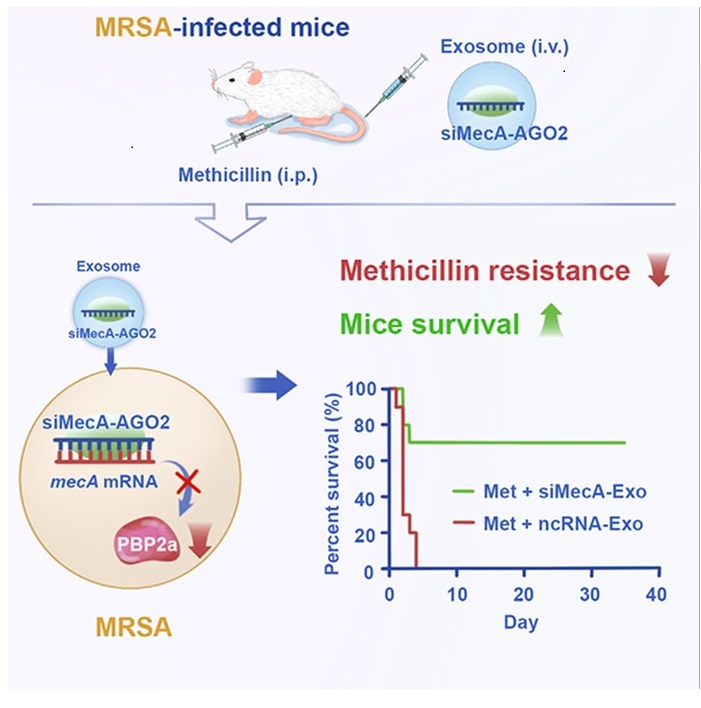
Moving beyond laboratory experiments, the researchers tested their approach in living animals, in particular BALB/c mice.
BALB/c mice, infected with lethal doses of MRSA, received daily treatments of methicillin combined with siMecA-Exos. The results were remarkable: 70% of mice survived when treated with the combination therapy, compared to almost complete mortality in control groups receiving methicillin alone or methicillin with control exosomes. Bacterial loads were significantly reduced in the blood, liver, spleen, and kidneys of treated mice. Inflammatory markers decreased, indicating reduced infection severity. Analysis of MRSA isolated from treated mice confirmed the presence of both siMecA and the AGO2 protein within the bacteria, along with reduced PBP2a expression, thereby verifying that the mechanism observed in laboratory experiments also operates within a living organism.
Innovation in delivery: self-assembled exosomes
Considering the potential challenges associated with exosome production for therapeutic use, the researchers developed an ingenious solution by leveraging the body’s own production abilities. They designed a genetic circuit that, when injected into mice, programs the animals’ liver cells to produce and release exosomes containing the desired siRNA.
This approach offers significant advantages: it eliminates complicated production and purification procedures, avoids potential immunogenicity and carcinogenicity concerns associated with foreign exosomes, and achieves biologically effective concentrations of siRNA in circulating exosomes. The results were compelling – mice infected with lethal doses of MRSA and treated with methicillin plus the siMecA genetic circuit showed a 90% survival rate over a 35-day observation period, a significantly reduced bacterial loads in blood and organs, and a consistent effectiveness against multiple MRSA strains.
Implications for the future of antibiotic therapy
This groundbreaking research opens several promising avenues for addressing antibiotic resistance. Unlike traditional antibiotics that require extensive chemical remodeling to overcome resistance mutations, the siRNA approach can be quickly adapted by simply changing the RNA sequence to match new target genes. The discovery also raises intriguing questions about whether mammals naturally use exosomes to regulate their microbiome through interspecies communication. Rather than replacing antibiotics, this approach could revitalize existing ones by silencing resistance mechanisms, potentially extending the useful life of our current antibiotic arsenal.
Challenges to overcome
Despite its promise, several limitations must be addressed before this technology can reach clinical application. The precise mechanism by which the siRNA-AGO2 complex inhibits bacterial protein translation requires further investigation. Current exosome purification methods may need refinement to ensure specificity. Moreover, studies across a broader range of bacterial strains and models are needed to generalize the findings. Lastly, the research focused on methicillin, which is rarely used today in the clinical practice, whereas further studies are needed to test the approach with currently used antibiotics.

Leave a Reply